Running AWS HealthOmics MCP Server on EC2
The Model Context Protocol (MCP) provides a standardized way for AI assistants to interact with external tools and data sources. AWS HealthOmics offers an MCP server that enables seamless integration with genomic workflows and data. This guide walks through setting up and running the AWS HealthOmics MCP server on an EC2 instance.
Prerequisites
Before getting started, ensure you have:
- AWS EC2 Instance: A running Ubuntu instance with appropriate IAM permissions for HealthOmics
- AWS CLI Configuration: Properly configured AWS credentials and region
- Python Environment: Python 3.8+ installed on the instance
Setup Process
1. Clone the MCP Repository
First, clone the AWS HealthOmics MCP server repository:
git clone https://github.com/awslabs/mcp.git
cd mcp/src/aws-healthomics-mcp-server
2. Install Dependencies
Install the required Python packages using the official installation method:
# Install using uvx (recommended)
uvx awslabs.aws-healthomics-mcp-server
# Or install from source
git clone https://github.com/awslabs/mcp.git
cd mcp/src/aws-healthomics-mcp-server
uv sync
uv run server.py
# Or install the package in development mode
pip install -e .
# Or install from PyPI
pip install awslabs-aws-healthomics-mcp-server
3. Configure AWS Credentials
This server requires AWS credentials with appropriate permissions for HealthOmics operations. Configure using:
# AWS CLI configuration
aws configure
# Enter your AWS Access Key ID
# Enter your AWS Secret Access Key
# Enter your default region (e.g., us-east-1)
# Enter your output format (json)
# Or use environment variables
export AWS_ACCESS_KEY_ID=your_access_key
export AWS_SECRET_ACCESS_KEY=your_secret_key
export AWS_REGION=us-east-1
# Or use IAM roles (recommended for EC2/Lambda)
# The instance will automatically use the attached IAM role
# Or use AWS profiles
export AWS_PROFILE=your_profile_name
Required IAM Permissions
The following IAM permissions are required for the HealthOmics MCP server:
{
"Version": "2012-10-17",
"Statement": [
{
"Effect": "Allow",
"Action": [
"omics:ListWorkflows",
"omics:CreateWorkflow",
"omics:GetWorkflow",
"omics:CreateWorkflowVersion",
"omics:ListWorkflowVersions",
"omics:StartRun",
"omics:ListRuns",
"omics:GetRun",
"omics:ListRunTasks",
"omics:GetRunTask",
"logs:DescribeLogGroups",
"logs:DescribeLogStreams",
"logs:GetLogEvents"
],
"Resource": "*"
}
]
}
4. Create Server Configuration
Create a server_config.json file with the following configuration:
{
"mcpServers": {
"aws-healthomics": {
"command": "python",
"args": ["-m", "awslabs.aws_healthomics_mcp_server.server"],
"cwd": "/home/ubuntu/mcp/src/aws-healthomics-mcp-server",
"env": {
"AWS_REGION": "us-east-1",
"AWS_PROFILE": "default",
"FASTMCP_LOG_LEVEL": "INFO"
}
}
}
}
This configuration specifies:
- The Python command to run the server
- The module path for the AWS HealthOmics MCP server
- The working directory
- Environment variables for AWS region, profile, and logging level
Environment Variables
AWS_REGION- AWS region for HealthOmics operations (default: us-east-1)AWS_PROFILE- AWS profile for authenticationFASTMCP_LOG_LEVEL- Server logging level (default: WARNING)
Running the MCP Server
1. Start the Server
Use the MCP CLI to start and manage the server:
uvx mcp-cli servers
This command will:
- Initialize the MCP CLI
- Start the AWS HealthOmics MCP server
- Display available servers and their status
2. Verify Server Status
Check if the server is running properly:
uvx mcp-cli ping
This will test the connection to the MCP server and display latency information.
3. List Available Tools
Explore the available tools provided by the HealthOmics MCP server:
uvx mcp-cli tools
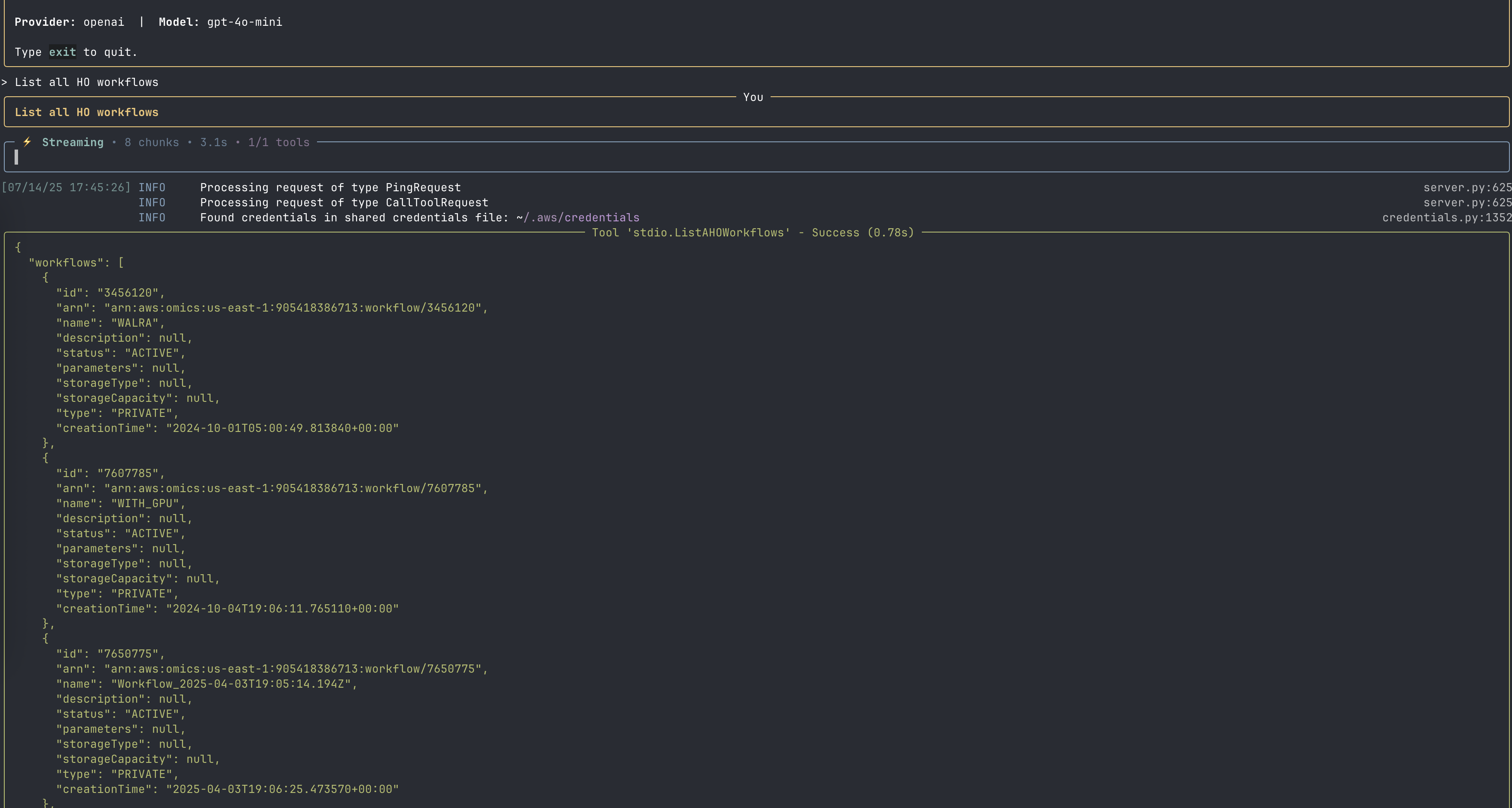
Available Tools
The AWS HealthOmics MCP server provides several tools for managing genomic workflows:
Workflow Management
- ListAHOWorkflows: List available HealthOmics workflows
- CreateAHOWorkflow: Create new HealthOmics workflows
- GetAHOWorkflow: Retrieve details about specific workflows
Run Management
- ListAHORuns: List workflow runs
- CreateAHORun: Start new workflow runs
- GetAHORun: Get details about specific runs
Storage Management
- ListAHOReferenceStores: List reference stores
- CreateAHOReferenceStore: Create new reference stores
- GetAHOReferenceStore: Get reference store details
Sequence Store Operations
- ListAHOSequenceStores: List sequence stores
- CreateAHOSequenceStore: Create new sequence stores
- GetAHOSequenceStore: Get sequence store details
Common Use Cases
Workflow Development
User: “Help me create a new genomic variant calling workflow”
- Use
CreateAHOWorkflowwith WDL/CWL/Nextflow definition - Package workflow files appropriately
- Validate syntax and parameters
Production Execution
User: “Run my alignment workflow on these FASTQ files”
- Use
StartAHORunwith appropriate parameters - Monitor with
ListAHORunsandGetAHORun - Track task progress with
ListAHORunTasks
Troubleshooting
User: “My workflow failed, what went wrong?”
- Use
DiagnoseAHORunFailurefor comprehensive analysis - Access specific logs based on failure type
- Provide actionable remediation steps
Performance Optimization
User: “How can I make my workflow run faster?”
- Use
AnalyzeAHORunPerformanceto identify bottlenecks - Review resource utilization patterns
- Suggest optimization strategies
Server Output Examples
When running the server, you’ll see output similar to:
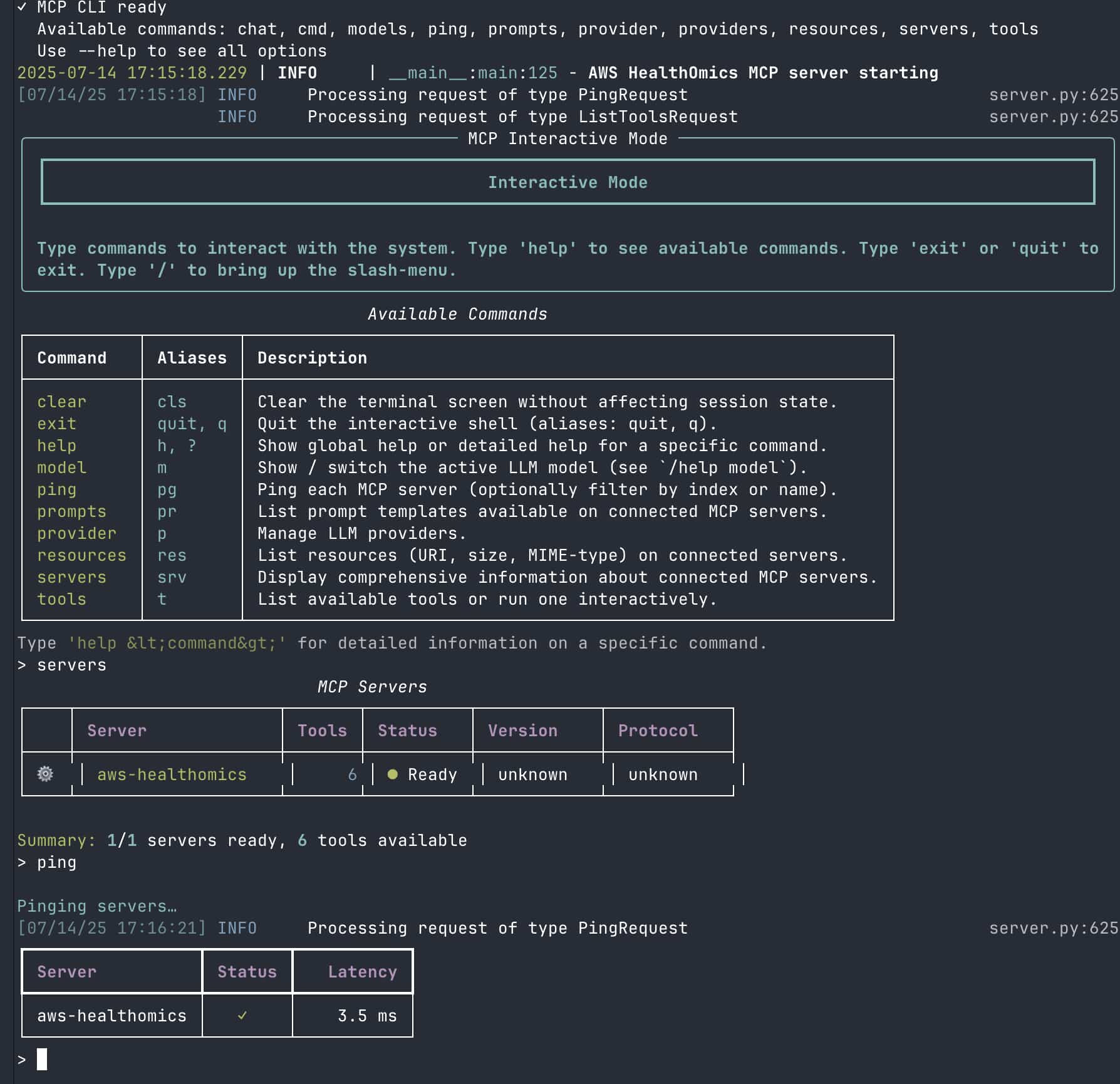
✓ MCP CLI ready
Available commands: chat, cmd, models, ping, prompts, provider, providers, resources, servers, tools
Use --help to see all options
2025-07-14 17:50:10.064 | INFO | __main__:main:125 - AWS HealthOmics MCP server starting
[07/14/25 17:50:10] INFO Processing request of type PingRequest
INFO Processing request of type ListToolsRequest
Server Status Check
┌────┬────────────────────┬───────┬──────────┬────────────┬────────────┐
│ │ Server │ Tools │ Status │ Version │ Protocol │
├────┼────────────────────┼───────┼──────────┼────────────┼────────────┤
│ * │ aws-healthomics │ 6 │ ● Ready │ unknown │ unknown │
└────┴────────────────────┴───────┴──────────┴────────────┴────────────┘
Summary: 1/1 servers ready, 6 tools available
Ping Test Results
┏━━━━━━━━━━━━━━━━━┳━━━━━━━━┳━━━━━━━━━━━┓
┃ Server ┃ Status ┃ Latency ┃
┡━━━━━━━━━━━━━━━━━╇━━━━━━━━╇━━━━━━━━━━━┩
│ aws-healthomics │ ✓ │ 3.0 ms │
└─────────────────┴────────┴───────────┘
Screenshots
Important Considerations
IAM Permissions
Ensure proper IAM roles with HealthOmics permissions as outlined in the configuration section above.
Regional Availability
Use GetAHOSupportedRegions to verify service availability in your desired AWS region.
Cost Management
Monitor storage and compute costs, especially with STATIC storage, as HealthOmics can be resource-intensive.
Data Security
Follow genomic data handling best practices and compliance requirements, including:
- HIPAA compliance for patient data
- Data encryption at rest and in transit
- Proper access controls and audit logging
Resource Limits
Be aware of service quotas and limits for concurrent runs:
- Maximum concurrent runs per account
- Storage limits for reference and sequence stores
- API rate limits for HealthOmics operations
Troubleshooting
Common Issues
- Authentication Errors: Ensure AWS credentials are properly configured
- Permission Denied: Verify IAM permissions for HealthOmics services
- Connection Timeouts: Check network connectivity and security groups
Debug Commands
# Check AWS credentials
aws sts get-caller-identity
# Test HealthOmics API access
aws omics list-workflows --region us-east-1
# Check server logs
tail -f /var/log/mcp-server.log
Conclusion
Setting up the AWS HealthOmics MCP server on EC2 provides a powerful way to integrate genomic workflows with AI assistants. The server offers comprehensive tools for managing workflows, runs, and data stores, making it easier to automate and orchestrate genomic analysis pipelines.
The MCP protocol standardizes the interaction between AI tools and external services, enabling more sophisticated automation and integration capabilities for genomic research and analysis workflows.